Mathematical biology is a branch of applied mathematics dealing with understanding and mathematically modelling the biological systems. The roots to this discipline stem from pioneering early works of Alan Turing who explained mathematically the structure of patterns such as cheetah spots, zebra stripes etc. in his seminal paper “The chemical basis of morphogenesis” using reaction diffusion systems. Ever since then, researchers have been looking into how biological systems work (from evolution, memory formation to self healing mechanisms) using a combination of several tools from non-linear dynamics, neural networks, signal processing and systems theory. We are interested in understanding the formation of both short term and long term memories using reaction diffusion principles inspired by pattern formation in animals. Our research has potential applications in engineering from search engines towards artificial biological systems.
Figure 1: The formation of spots on Cheetah can be explained using reaction-diffusion equations. [Courtesy: Wikipedia]
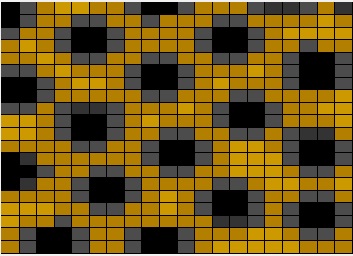
Figure 2: Cheetah spots generated using reaction-diffusion equations in MATLAB.
It is amazing to see how human brain gathers inputs from multiple sources and processes information parallely even in an unknown environment. This is possible because we have memory, categorized as 1) short term memory and 2) long term memory and brain does self organization in a unique way from over thousand different neuronal types and associated connections. Whenever an input is received by the human brain, certain neurons get excited and the information about the input is spread across the system of neurons. This spread of information can be thought of as a wave propagating over the space and time forming an associative spatio-temporal memory. At PNSIL, we are trying to understand and mathematically model spatio-temporal memories from first principles based on Turing’s reaction-diffusion equations and applications beyond this.
Journals
- P. Gowgi, A. Machireddy and S. S. Garani, “Spatio-temporal Memories for Missing Samples Reconstruction”, in IEEE Trans. on Neural Netw. and Learning Syst. (doi: 10.1109/TNNLS.2021.3062463).
- P. Gowgi and S. S. Garani, “Temporal Self-Organization: A Reaction-Diffusion Framework for Spatiotemporal Memories”, in IEEE Trans. on Neural Netw. and Learning Syst., vol. 30, no. 2, pp. 427-448, Feb. 2019.
- J. C. Principe, N. R. Euliano and S. Garani, “Principles and networks for self organization in space time”, Special Issue of Journal of Neural Networks, Elsevier Press, vol. 15, pp. 1069-1083, Oct. 2002.
Book chapters
- S. Garani and J. C. Principe, “Dynamic vector quantization of speech,” Proc.of Workshop on Self Organizing Maps., Springer-Verlag: London, pp. 238-245, July 2001.
Conferences
- A. Machireddy, P. Gowgi and S. S. Garani, “Extracting Temporal Correlations Using Hierarchical Spatio-Temporal Feature Maps”, in IEEE Int. Joint Conf. on Neural Networks (IJCNN) (accepted).
- P. Gowgi and S. S. Garani, “Hessian-based Bounds on Learning Rate for Gradient Descent Algorithms”, in IEEE Int. Joint Conf. on Neural Networks (IJCNN), Glasgow, UK, July 2020.
- A. Machireddy and S. S. Garani, “Guessing the Code: Learning Encoding Mappings Using the Back-Propagation Algorithm”, in IEEE Int. Joint Conf. on Neural Networks (IJCNN), Budapest, Hungary, July 2019.
- P. Gowgi, A. Machireddy and S. S. Garani, “Priority-based Soft Vector Quantization Feature Maps”, in IEEE. Int. Joint Conf. on Neural Networks (IJCNN), Rio, Brazil, July 2018.
- A. Machireddy and S. S. Garani, “Data Dependent Adaptive Prediction and Classification of Video Sequences”, in Int. Conf. on Artificial Intelligence and Soft Computing (ICAISC), Zakopane, Poland, June 2018.
- P. Gowgi and S. S. Garani, “Density Transformation and Parameter Estimation from Back Propagation Algorithm”, in IEEE. Int. Joint Conf. on Neural Networks (IJCNN), Vancouver, Canada, July 2016.
- P. Gowgi and S. G. Srinivasa, “Spatio-temporal map formation based on a Potential Function”, in IEEE. Int. Joint Conf. on Neural Networks (IJCNN), Killarney, Ireland, July 2015.
- S. Garani and J. C. Principe, “A spatio-temporal vector quantizer for missing samples reconstruction,” IEEE. Proc. Int. Joint Conf. on Neural Networks., vol. 4, pp. 2913-2917, June 2001.